Structures considered key to gene expression are surprisingly fleeting
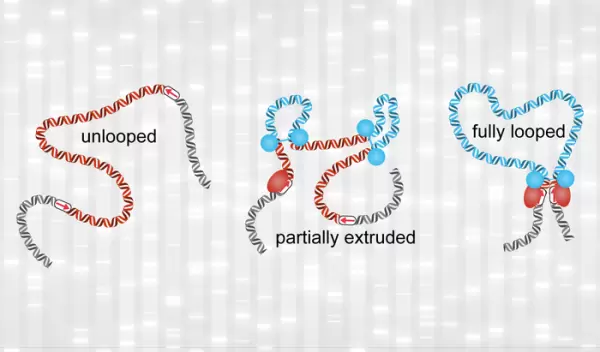
In human chromosomes, DNA is coated by proteins, forming a long-beaded "string." This string is folded into numerous loops, which are believed to help cells control gene expression and facilitate DNA repair, among other functions. A study by MIT scientists suggests that these loops are very dynamic and shorter-lived than previously thought.
The U.S. National Science Foundation-supported researchers monitored the movement of one stretch of the genome in a living cell for about two hours. They saw that this stretch was fully looped for only 3% to 6% of the time, with the loop lasting for only about 10 to 30 minutes. The findings suggest that scientists' current understanding of how loops influence gene expression may need to be revised, the researchers say.
"Many models in the field have been these pictures of static loops regulating processes, but what our new paper shows is that this picture is not correct," said Anders Sejr Hansen at MIT. "We suggest that the functional state of these domains is much more dynamic."
Hansen is one of the senior authors of the new study, along with Leonid Mirny at MIT and Christoph Zechner at the Max Planck Institute of Molecular Cell Biology and Genetics in Dresden, Germany. MIT researchers Michele Gabriele and Simon Grosse-Holz are the lead authors of the paper, which appears in Science.
Using computer simulations and experimental data, scientists have shown that loops in the genome are formed by a process called extrusion, in which a molecular motor promotes the growth of progressively larger loops. The motor stops each time it encounters a "stop sign" on DNA. The motor that extrudes such loops is a protein complex called cohesin, while the DNA-bound protein CTCF serves as the stop sign. These cohesin-mediated loops between CTCF sites were seen in previous experiments.
However, those experiments only offered a snapshot of a moment in time, with no information on how the loops change over time. In the new study, the researchers developed techniques that allowed them to fluorescently label CTCF DNA sites so they could image the DNA loops over several hours. They also created a new computational method that can infer the looping events from the imaging data.
According to Hansen, if chromatin loops are so short-lived, new models may be needed to understand how the 3D structure of the genome regulates gene expression, DNA repair and many other cellular processes. Manju Hingorani, a program director in NSF's Division of Molecular and Cellular Biosciences, added that "the findings are an example of how static views of macromolecules come to life and reveal their secret workings, as live-cell imaging technologies achieve finer spatial and temporal resolution."


